Finding new drugs to fight bad bugs
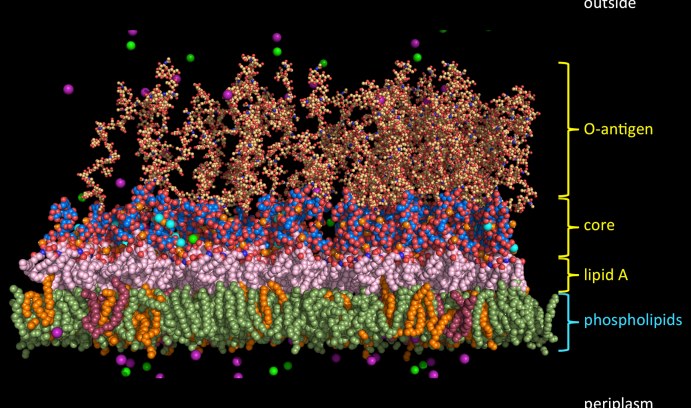
Gram-negative bacterial outer membrane molecular complexity: An illustration of a typical E. coli outer membrane is shown, along with the molecular system used to represent the complexity in molecular dynamics simulations. (Image courtesy of Wonpil Im)
As bacteria change their behavior to foil the drugs designed to kill them, illnesses that were once relatively easy to treat become difficult to cure. The phenomenon is known as antibiotic resistance, and unless it is overcome, a UK-funded analysis warns, drug-resistant bacterial infections could one day kill more people worldwide than cancer.
Progress in developing “new drugs for bad bugs” (a phrase coined by the European Commission’s Innovative Medicines Initiative) depends on information—especially when it comes to translocation, the action an antibiotic must take to penetrate the outer membrane of a bacterial cell in order to reach and destroy its target.
The question then becomes: Which drugs are able to penetrate the outer membranes of bacteria, and which are not?
That, according to Wonpil Im, is the mystery. Solving it is especially important when it comes to the “Big Bads” of bacterial infections known as Gram-negative pathogens like “superbugs.” These bacterial species are resistant to multiple drugs and are increasingly resistant to most available antibiotics. The infections they cause can be lethal. Some strains of the Gram-negative pathogen E. coli can cause serious food poisoning, for example, while certain strains of the Gram-negative bacteria called Vibrio cholera cause cholera, an infectious, often fatal disease of the small intestine.
Im, professor of biological sciences and bioengineering and Presidential Endowed Chair in Health Science and Engineering at Lehigh, is an expert in molecular simulation systems. His CHARMM-GUI, a web-based graphical user interface that he helped design to model complex biomolecular systems, aids in simulation design to produce models that usually require considerable experience and time (as much as several weeks) in just a few minutes or hours.
Now, through modeling and simulation, Im and his team have revealed the bilayer properties of 21 distinct Lipid A types from 12 Gram-negative bacterial species. Im and his colleagues from the University of Kansas, University of Maryland and Stockholm University investigate the differences and similarities of the membrane properties such as area per lipid, hydrophobic thickness, and acyl chain order. The group also examined the influence of several neutralizing ion types (Ca2+, K+, and Na+) on bacterial membrane properties, including lipid diffusion coefficients, ion residence times, and compressibility modulus that are experimentally comparable. Their results were published yesterday (Oct. 18) in an article in Biophysical Journal titled “Bilayer Properties of Lipid A from Various Gram-negative Bacteria.”
Lipid A is one of three regions that make up lipopolysaccharides (LPS), an integral component of the outer leaflet of the outer membrane of Gram-negative bacteria where they shield the bacterium from environmental threats. Because the LPS make the outer membrane more impermeable to antibiotics, understanding and manipulating LPS membrane stability could be an effective way to develop new Gram-negative antibiotics. Lipid A is the anchoring region of LPS and is responsible for Toxic Shock Syndrome and sepsis.
“The more information we can provide about the properties of the outer membrane of these so-called ‘bad bugs,’” says Im, “the quicker researchers can determine which compounds might penetrate and effectively target a specific protein—even turn it off.”
From the study: “…the resulting characteristics of different Lipid A obtained in this study form a basis toward the modeling and simulation of the outer membranes (with and without outer membrane proteins) of various Gram-negative bacteria. In particular, all Lipid A models in this study are available through LPS Modeler in CHARMM-GUI, so that these models can be used to further our understanding of the structure, dynamics, and function of various bacterial outer membranes.”
“My dream,” says Im, “is that one day pharmaceutical companies could come to my lab with 100 compounds with the potential to kill a given Gram-negative bacteria and ask: Which ones should we try? And we could tell them.”
To advance this notion, Im is organizing a “Workshop to Take Aim at Bacteria” at Lehigh on Friday, Nov. 18. The workshop is designed to bring experts in research and policy (including a leader in the Innovative Medicines Initiative) to help speed antibiotic drug development.
Story by Lori Friedman
Posted on:




